Debugging in R
Recently I’ve been part of beta testing a new R package. The package allows users to apply different masks to spcies distribution models to create more ecologically realistic range models. The package itself is very cool, but since it hasn’t been released yet, this post will be short on ecology. Instead, I’m going to focus on the process I’ve been using to debug the package. This debugging thought process and the different functions that make it possible are things I wish I’d been formally taught while learning R (but better late than never!).
traceback
The traceback() function is definitely one of those that I wished I learned sooner. Basically, traceback() shows you the series of functions called, including the one resulting in the last error. For example, when I ran traceback() after the function I was testing threw an error, I got the following readout:
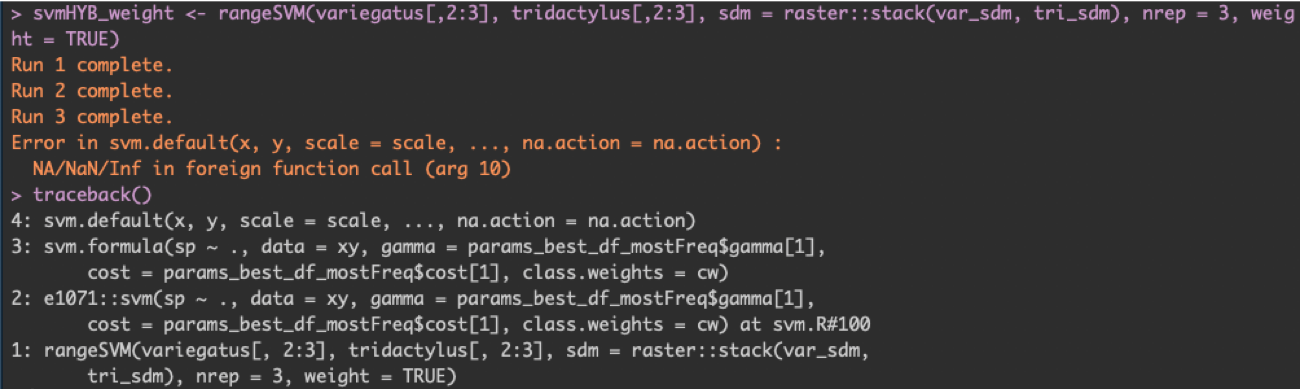
When I tried to use the function rangeSVM(), I got an error, as we can see in the first line of code I ran. By running traceback(), I can see the order of functions that rangeSVM() uses: first, it called the svm() function from the R package e1071, which in turn called svm.formula(), which finally called svm.default(). This last function, svm.default() is what produced the error (as we can see from the error message, as well).
Running traceback() can be especially helpful in situations like this, where the error message comes from a function called internally by the function you actually ran. Here, the error does not come from the rangeSVM() function, so reading the documentation for rangeSVM() could not save me. Now I know that the error actually comes from a function in the e1071 package, so I could check out the source code to figure out what kind of issue causes that error message.
Checking source code
There is a CRAN GitHub account that is an unofficial read-only mirror of all CRAN packages. Typically, when I want to read through the code for a particular function in a package, this is what I use. To dig a little deeper into what was going wrong with the svm() function, I found the code here. This allowed me to look at the function definitions for svm.formula() as well as svm.default().
debug
Reading through the source code is helpful, but wouldn’t it be even better if you could watch R step through the function line by line until the error? Fortunately the handy debug() function does exactly that. By running the following:
debug(rangeSVM)
svmHYB_weight <- rangeSVM(variegatus[,2:3], tridactylus[,2:3], sdm = raster::stack(var_sdm, tri_sdm), nrep = 3, weight = TRUE)I could see that the error occurred at line 100 of svm.R:
# Browse[2]>
# debug at /Users/hellenfellows/OneDrive - AMNH/Wallace/maskRangerBetaTesting/maskRangeR/R/svm.R#100: m <- e1071::svm(sp ~ ., data = xy, gamma = params_best_df_mostFreq$gamma[1],
# cost = params_best_df_mostFreq$cost[1], class.weights = cw)
# Browse[2]>
# Error in svm.default(x, y, scale = scale, ..., na.action = na.action) :
# NA/NaN/Inf in foreign function call (arg 10)The Browse> prompt at the beginning of the lines indicate that the debugger is working. The debug at part shows the next line of code to be executed when you hit enter next. R will continue to show you the following line of code as you press enter (up until you hit the error). If you are running all of this in RStudio, it will also conveniently highlight the lines in the source code that will be run next. Also, when you want to get out of the debugger so you can run that function again later without debugging, run undebug(rangeSVM), for example.
My initial suspicion was that the function was failing during one of the runs of a for loop, however the debugger showed me that the function was making it through all the runs of the for loop. Instead, the error occurred during the final step of rangeSVM() when the function attempted to integrate the results of all runs of the for loop to create a final support vector machine model.
get
To figure out why this problem was occurring, I wanted to be able to see the values of the different variables passeed to that final step of the function. Handily enough, while in the debugger, you can use the function get("variable") to check the status of the different variables (the name of the variable needs to be in quotation marks).
For example:
debug(rangeSVM)
svmHYB <- rangeSVM(variegatus[,2:3], tridactylus[,2:3], sdm = raster::stack(var_sdm, tri_sdm), nrep = 3)
# Browse[2]>
# debug at /Users/hellenfellows/OneDrive - AMNH/Wallace/maskRangerBetaTesting/maskRangeR/R/svm.R#96: params_best_df$params <- paste0(params_best_df$gamma, params_best_df$cost)
# Browse[2]> get("params_best_df")
# gamma cost class.weights
# 38 0.5000000 2 1
# 85 0.0078125 2048 1
# 95 0.0078125 8192 1This was the final step of the process that actually allowed me to figure out the problem:
# Browse[2]> get("params_best_df_mostFreq")
# [1] gamma cost
# <0 rows> (or 0-length row.names)Without going into too much detail about the function itself, I was able to tell that the parameters I was feeding into the support vector machine function somehow didn’t exist: the variable params_best_df_mostFreq had 0 rows.
Other tips and tricks
I got much of these resources from a very helpful blog post called “Tracking down errors in R” by Pete Werner (also available as a post on R-bloggers). This post goes through a slightly simpler example of debugging which is reproducible (something I have not attempted to do here), so it is very helpful for seeing the process on a simpler function. In addition to the techniques I used, Pete also explains how you can turn warning messages into errors in the case that your function is throwing a troubling warning message that you suspect is causing an error later on.